Note
Click here to download the full example code
Finding a discrete maximum entropy distribution
Here we consider a simple convex optimization problem to illustrate how to use Cooper. This example is inspired by this StackExchange question:
I am trying to solve the following problem using Pytorch: given a 6-sided die whose average roll is known to be 4.5, what is the maximum entropy distribution for the faces?
14 import copy
15 import os
16
17 import matplotlib.pyplot as plt
18 import numpy as np
19 import style_utils
20 import torch
21
22 import cooper
23
24 torch.manual_seed(0)
25 np.random.seed(0)
26
27
28 class MaximumEntropy(cooper.ConstrainedMinimizationProblem):
29 def __init__(self, mean_constraint):
30 self.mean_constraint = mean_constraint
31 super().__init__(is_constrained=True)
32
33 def closure(self, probs):
34 # Verify domain of definition of the functions
35 assert torch.all(probs >= 0)
36
37 # Negative signed removed since we want to *maximize* the entropy
38 neg_entropy = torch.sum(probs * torch.log(probs))
39
40 # Entries of p >= 0 (equiv. -p <= 0)
41 ineq_defect = -probs
42
43 # Equality constraints for proper normalization and mean constraint
44 mean = torch.sum(torch.tensor(range(1, len(probs) + 1)) * probs)
45 eq_defect = torch.stack([torch.sum(probs) - 1, mean - self.mean_constraint])
46
47 return cooper.CMPState(
48 loss=neg_entropy, eq_defect=eq_defect, ineq_defect=ineq_defect
49 )
50
51
52 # Define the problem and formulation
53 cmp = MaximumEntropy(mean_constraint=4.5)
54 formulation = cooper.LagrangianFormulation(cmp)
55
56 # Define the primal parameters and optimizer
57 rand_init = torch.rand(6) # Use a 6-sided die
58 probs = torch.nn.Parameter(rand_init / sum(rand_init))
59 primal_optimizer = cooper.optim.ExtraSGD([probs], lr=3e-2, momentum=0.7)
60
61 # Define the dual optimizer. Note that this optimizer has NOT been fully instantiated
62 # yet. Cooper takes care of this, once it has initialized the formulation state.
63 dual_optimizer = cooper.optim.partial_optimizer(
64 cooper.optim.ExtraSGD, lr=9e-3, momentum=0.7
65 )
66
67 # Wrap the formulation and both optimizers inside a ConstrainedOptimizer
68 coop = cooper.ConstrainedOptimizer(formulation, primal_optimizer, dual_optimizer)
69
70 state_history = cooper.StateLogger(save_metrics=["loss", "eq_defect", "eq_multipliers"])
71
72 # Here is the actual training loop
73 for iter_num in range(5000):
74 coop.zero_grad()
75 lagrangian = formulation.composite_objective(cmp.closure, probs)
76 formulation.custom_backward(lagrangian)
77 coop.step(cmp.closure, probs)
78
79 # Store optimization metrics at each step
80 partial_dict = {"params": copy.deepcopy(probs.data)}
81 state_history.store_metrics(formulation, iter_num, partial_dict)
82
83 all_metrics = state_history.unpack_stored_metrics()
84
85 # Theoretical solution
86 optim_prob = torch.tensor([0.05435, 0.07877, 0.1142, 0.1654, 0.2398, 0.3475])
87 optim_neg_entropy = torch.sum(optim_prob * torch.log(optim_prob))
88
89 # Generate plots
90 fig, (ax0, ax1, ax2) = plt.subplots(nrows=1, ncols=3, sharex=True, figsize=(15, 3))
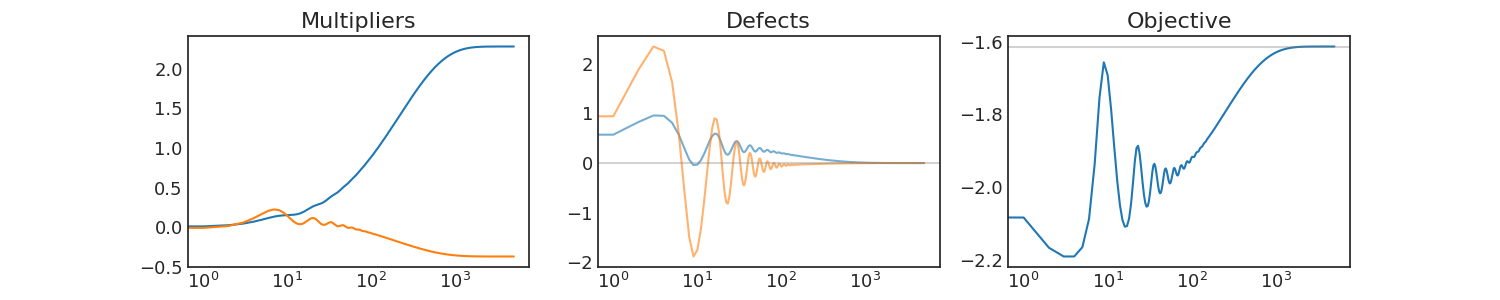
91
92 ax0.plot(all_metrics["iters"], np.stack(all_metrics["eq_multipliers"]))
93 ax0.set_title("Multipliers")
94
95 ax1.plot(all_metrics["iters"], np.stack(all_metrics["eq_defect"]), alpha=0.6)
96 # Show that defect remains below/at zero
97 ax1.axhline(0.0, c="gray", alpha=0.35)
98 ax1.set_title("Defects")
99
100 ax2.plot(all_metrics["iters"], all_metrics["loss"])
101 # Show optimal entropy is achieved
102 ax2.axhline(optim_neg_entropy, c="gray", alpha=0.35)
103 ax2.set_title("Objective")
104
105 [_.semilogx() for _ in (ax0, ax1, ax2)]
106 plt.show()
Total running time of the script: ( 0 minutes 9.553 seconds)